Abstract
Current prognostic models of gene expression profiles for acute myeloid leukemia (AML) are inconsistent at predicting clinical outcomes for individual patients. None include data on T-cell function-related genes (TFGs). Here, we identified 218 candidate genes from the bone marrow mononuclear cells bulk RNA-seq matrix in the SYSUCC-1 cohort based on the immune ProcartaPlex panel. We then identified 83 genes as TFGs with a significant Spearman correlation to a T-cell subset fraction or PD-1/CTLA-4 MFI expression.
Next, we interrogated data of RNA matrices from 1,611 persons with AML extracted from public databases arrayed in training and 3 cohorts. Using a random survival forest variable hunting algorithm we developed an 8 gene T-cell function-related signature which correlated with survival after adjusting for other prognostic co-variates. The final TFG signature was determined based on validation with the best predictive performance in the real world dataset using the formula:
TFG risk scores = (-0.177 × expression of CD27) + (0.26 ×expression of IL2RA) -(0.19 × expression of CD96) - (0.098 × expression of TIMP1) + (0.19 × COL18A1 expression of COL18A1) - (0.017 × expression of HGF) - (0.196 × GDF15 expression of GDF15) - (0.161 × CLEC11A expression of CLEC11A).
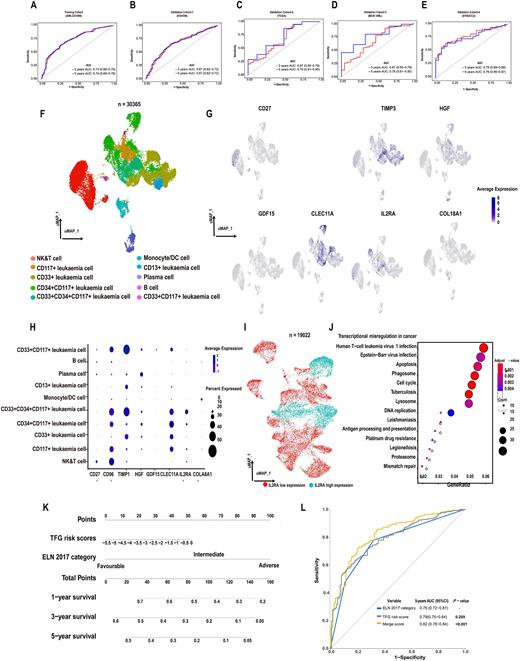
The model had robust predictive accuracy in the training and 4 external validation cohorts with 5-year areas under receiver-operator characteristic curve (AUROCs) of 0.67-0.76(Figure 1A-D). The signature was divided into high- and low-risk scores using optimum cutoff value. 5-year survivals in the 4 high-risk cohorts were 6-23% compared with 42-58% in the 3 low-risk cohorts (all p-values < 0.001). In multi-variable analyses a high-risk score independently predicted briefer survival with Hazard Ratios (HR) = 1.28-2.59 in the training and validation cohorts (all p-values < 0.05). Gene set enrichment analyses (GESA) indicated significant enrichment for genes involved in immune suppression pathways in the high-risk cohorts. Accuracy of gene identification was validated in a real world cohort of 88 subjects by determining cognate plasma protein concentrations of these genes (Figure 1E).
To investigate TFG expression depending on cell type and function in the AML bone marrow microenvironment, we performed single-cell analysis of 36,477 cells from 16 subjects with de novo AML. Using a UMAP analysis, 36,477 bone marrow-derived cells were segregated into 10 populations according to the corresponding cell markers (Figure 1F). The expression profile of model TFGs showed that most of the genes, including CD96, TIMP1, HGF, CLEC11A, and IL2RA, were upregulated in leukaemia cells. In addition, CD27 and CD96 were highly upregulated in NK&T cells (Figure 1G-H). To further understand the role of IL2RA (the gene with the largest weight)-driven functional changes in leukaemia cells of AML patients, we divided all leukaemia cells into two groups according to the expression level of IL2RA (IL2RA high- and low-expression groups) and performed DEG (Differential gene) analysis (Figure 1I). The KEGG pathways enriched in the upregulated DEG were mainly specific to transcriptional dysregulation in cancer and associated with cancer progression and immune-related responses, including human T-cell leukaemia virus-1, Epstein-Barr virus (EBV) infection and tuberculosis pathways (Figure 1J).
Finally, the TFG signature and ELN2017 risk cohorts were integrated to generate a model using a nomogram, which had an accurate predictive value for 1-, 3-, and 5-year survival (Figure 1K) and increased the AUROC value from 0.76 (0.72, 0.81) to 0.82 (0.78, 0.84; P < 0.001; Figure 1L).
In summary, we developed a survival prediction score based on the expression of T-cell function related genes. Adding our score to the 2017 ELN risk score increased survival prediction accuracy. The combined nomogram may help physicians select appropriate therapies.Detection of plasma protein concentrations in peripheral blood may be a simple and effective method for predicting the survival outcome and prognosis of patients with AML.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.